Cerebral small vessel disease is a leading cause of stroke and dementia for which no mechanism-based drugs are currently available. The first genomic study on perivascular space burden (PVS), an emerging brain imaging marker of cerebral small vessel disease, revealed 24 genetic risk loci for PVS in a large meta-analysis of genome-wide association studies (N>40,000), mostly in the white matter. Importantly, most white matter PVS loci reached nominally significant associations with PVS in at least one of two smaller follow-up cohorts, in young adults and in older East-Asian adults (i-Share, Nagahama), supporting extension of these findings across the lifespan and across ancestries. We found significant enrichment of PVS loci in genes involved in early-onset leukodystrophies and expressed in fetal brain vascular endothelial cells, supporting involvement of developmental processes. Transcriptome-wide association studies further reveal 12 genes to prioritize for functional follow-up, including targets of drugs investigated for vascular and cognitive disorders. These findings provide entirely novel insights into the biology and clinical significance of PVS and open new avenues for therapeutic developments.
The results of the first genomic study on PVS were published in the journal Nature Medicine today, 17 April 2023. The study was based on DNA samples of more than 40,000 participants, of whom more than 9,000 had extensive PVS burden. Participants were of European, Hispanic, East-Asian, African-American ancestry. They were derived from numerous population-based cohorts and biobanks.
The study was conducted by members of the NeuroCHARGE consortium, involving numerous international consortia and networks, such as BRIDGET and ISGC, and investigators from 12 countries. It was co-led by three research centers at Bordeaux Population Health research center (University of Bordeaux, France), the Latin American Brain Health (BrainLat) institute at Universidad Adolfo Ibáñez in Santiago (Chile), and Erasmus MC University Medical Center in Rotterdam (The Netherlands), together with research departments at the Biggs Institute, UT Health San Antonio (USA), University of Edinburgh (UK), and Kyoto University Graduate School of Medicine (Japan).
This study provides unprecedented insight into the biology underlying PVS and converging evidence that mechanisms leading to cerebral small vessel disease may find their roots much earlier in life than previously thought. This has important implications for prevention strategies of cerebral small vessel disease and related stroke and dementia.
Stéphanie Debette, SHIVA coordinator and one of the corresponding authors of this publication
Importantly, AI and machine-learning approaches, very recently developed by different members of our consortium, were gamechangers in this endeavor, enabling us to rapidly and reliably assess PVS burden in large and diverse datasets,
Hieab Adams, Professor of Genetic Diversity jointly at BrainLat, Chile, and the Erasmus MC, the Netherlands, another corresponding author of the study.
More than 95% of research participants in genomic research on imaging markers of brain aging are of European ancestry. The expansion of this project to the Nagahama study, a large Japanese population-based cohort, is an important step forward in enhancing cross-ancestry contributions to brain aging genomics research.
Fumihiko Matsuda, Professor and Director of the Center for Genomic Medicine at Kyoto University Graduate School of Medicine (Japan), another leader of the study.
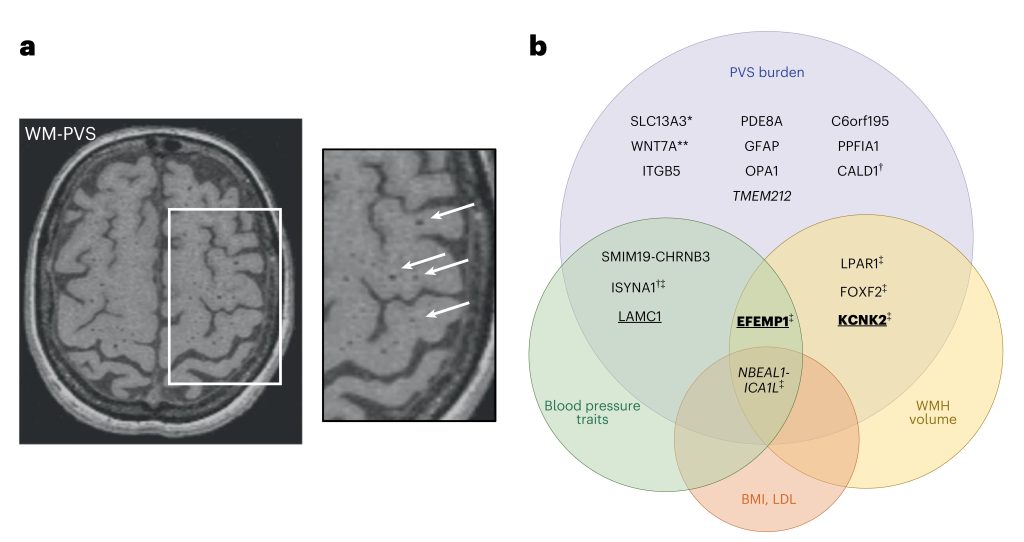
PVS are physiological spaces surrounding small vessel walls, and high PVS burden is thought to reflect impairment of brain fluid and waste clearance. PVS burden is highly heritable, but its genetic underpinnings were unknown so far. This meta-analysis has been conducted within a large international collaborative consortium, and includes 22 population-based cohorts. Given differential associations with risk factors and neurological traits and anatomical differences, the authors ran analyses separately for PVS in the white matter, basal ganglia and hippocampus. Interestingly 2/3 of PVS risk loci point to pathways that were not mediated by established risk factors, involving extra-cellular matrix, blood-brain barrier, membrane transport, and vascular development. The researchers found differential patterns of shared genetic variation with blood pressure and neurological traits according to PVS location, the strongest evidence for causal associations between high blood pressure and PVS and between PVS burden and stroke being observed for PVS in basal ganglia and, to a lesser extent, PVS in hippocampus.
These findings provide new insight into the biology of PVS across the adult lifespan and its contribution to the pathophysiology of cerebral small vessel disease, a major cause of stroke and dementia worldwide, with potential for genetically informed prioritization of drug targets for prevention trials.
Marie-Gabrielle Duperron, SHIVA physician and researcher, fist author of this publication
With increasing development of AI-based computational methods for PVS quantification and characterization, future genomic studies will enable more granular explorations, e.g. of genetic risk variants for PVS volume, width, shape, etc. They will also allow to study the genomics of MRI-markers of cerebral small vessel disease in a more integrated way and to derive disease patterns with specific molecular signatures and clinical correlates.
Marie-Gabrielle Duperron, SHIVA physician and researcher, fist author of this publication
Original publication:
Genomics of perivascular space burden unravels early mechanisms of cerebral small vessel disease (2023).
Marie-Gabrielle Duperron, Maria J. Knol, Quentin Le Grand, Tavia E. Evans, Aniket Mishra, Ami Tsuchida, et al. Nature Medicine. doi: 10.1038/s41591-023-02268-w
More information:
Pr. Stephanie Debette
University of Bordeaux, Inserm, Bordeaux Population Health Research Center, UMR 1219
Department of Neurology, Bordeaux University Hospital
146, rue Léo Saignat, 33076 Bordeaux, France
T: +33 (0)5 57 57 16 59; C: +33 (0)6 20 89 62 24
E-mail: stephanie.debette@u-bordeaux.fr
Dr. Hieab H. H. Adams
Department of Clinical Genetics; Department of Radiology and Nuclear Medicine
Erasmus MC University Medical Center; Wytemaweg 80, 3015 CE, Rotterdam, the Netherlands
Latin American Brain Health (BrainLat), Universidad Adolfo Ibáñez, Santiago, Chile
T: +31 10 70 33559; Fax number: +31 10 70 43489
E-mail: h.adams@erasmusmc.nl